The role of the 3' UTR in gene regulation

Untranslated regions comprise more than 40% of the average transcript (Ensembl, 2020). They are more conserved between species, and more constrained within humans, than ENCODE regulatory regions (enhancers). They perform a large range of functions, including localization of mRNAs, regulating protein-protein interactions, and importantly, regulating transcript stability and translation efficiency. Despite this they are less well studied than both transcriptional regulatory regions (promoters and enhancers) and non-coding RNAs.
Understanding of 3’ UTRs has medical and translational implications. Several diseases are affected by mutations in UTRs including Muscular Atrophy, Lupus, and several types of cancer. UTRs are also important from a synthetic biology standpoint. The dynamics of protein production in mRNA-based therapeutics depends on the stability of the transcript and the efficiency with which it is translated. This, in turn, is to a large extent determined by the sequence of the UTRs. Thus, it is critically important that therapeutic mRNA have optimized UTRs.
The function of 3' UTR introns
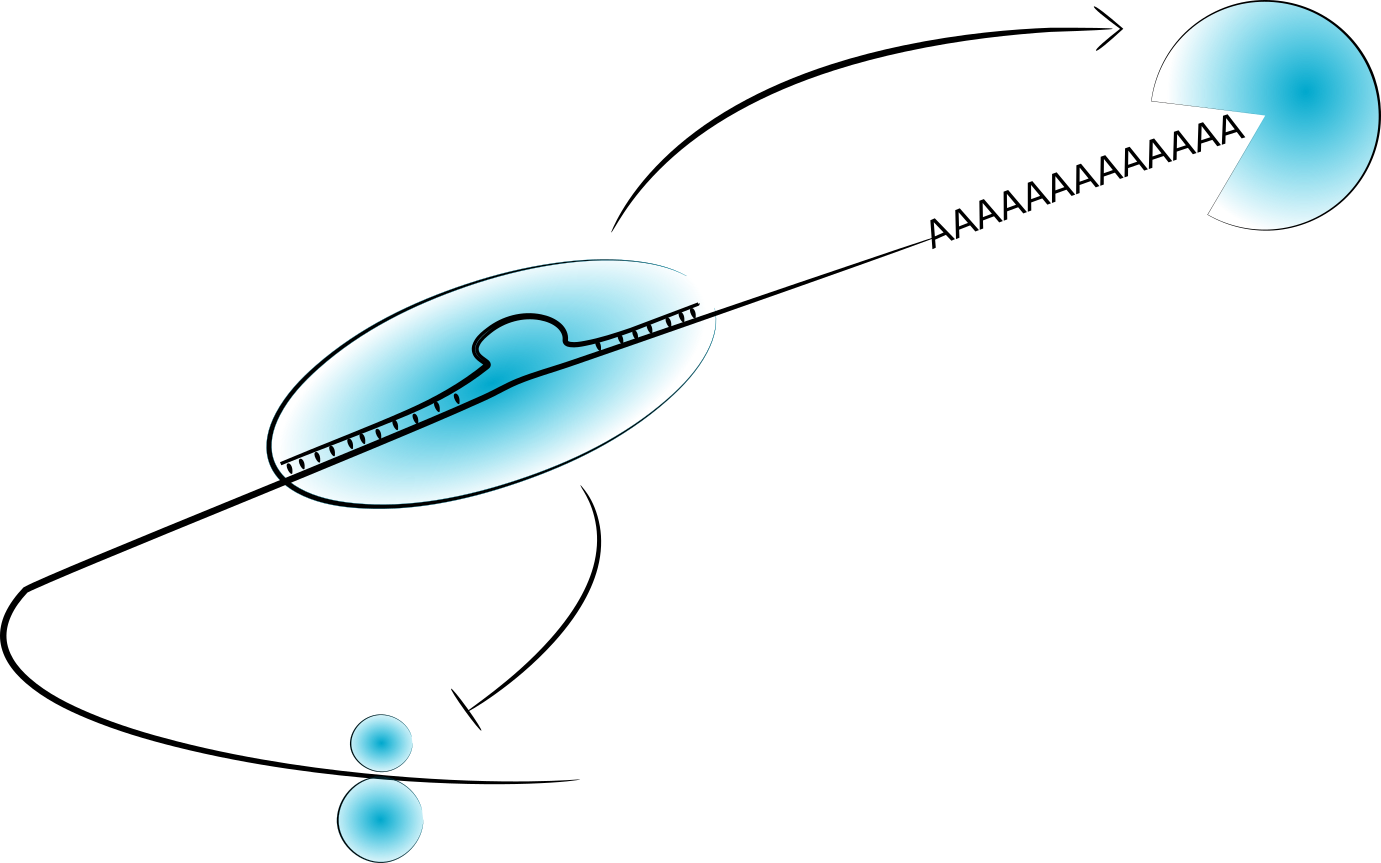
Shortening of UTRs in highly proliferative cell-types, such as hematopoietic and cancer, is a well established non-genetic mechanism by which the sequence content of 3’ UTRs vary. Such shortening is thought to remove regulatory sequences and stabilize transcripts. An alternative way in which 3’ UTR sequence might vary is via the inclusion or exclusion of alternative spliced introns in the 3’ UTR. The splicing of such introns would be expected to trigger nonsense mediated decay (NMD), destabilising transcripts. However, as part of a BBSRC funded project we have identified thousands of examples of highly expressed transcripts with introns in their 3’ UTRs in both cancer patient samples and healthy controls, and stem-cell lines. In many cases these introns overlap potential regulatory sequences. We are investigating the consequences of these using a mixture of high-throughput RNA stability measurements and reporter gene assays.
Synthetic UTRs
We have developed pipelines for generating synthetic 3’ UTRs (SUTRs) using sequence motifs associated with stability/instability or high/low translation efficiency. We have generated a library of SUTRs built from motifs we predict to stabilise transcripts, which we will submit to massively parallel assays. We also hope to use the data to understand something of the regulatory grammar of 3’ UTRs. Is the effect of motifs additive or multiplicative? How does the effect scale with multiple copies of the same motif? Are orders, spacing, and location important? We will analyse this with a range of different statistical modelling/machine learning approaches. We are also working with industrial partners to use this approach to generate optimized sequences for applied uses, including mRNA therapeutics.